Sample name: K562_RNA-seq_20
Looper stats summary
| sample_name |
K562_RNA-seq_20 |
| sample_desc |
80% K562 PRO-seq + 20% K562 RNA-seq |
| treatment |
none |
| replicate |
1 |
| toggle |
1 |
| protocol |
PRO |
| organism |
human |
| read_type |
SINGLE |
| cell_type |
K562 |
| purpose |
mRNA contamination; FRiF/PRiF |
| umi_status |
FALSE |
| umi_length |
0 |
| data_source |
SRA |
| read1 |
/project/shefflab/data/guertin/fastq/K562_20pct_RNArc_r2.fastq.gz |
| srr |
K562_20pct_RNArc_r2 |
| Sample_title |
K562 RNA spike-in 20pct RC |
| Sample_geo_accession |
GSM1480327;GSM765405 |
| Sample_source_name_ch1 |
K562 cells |
| File_mb |
806.51 |
| Read_type |
single |
| Genome |
hg38 |
| Raw_reads |
10000000 |
| Fastq_reads |
10000000 |
| Reads_with_adapter |
7511497.0 |
| Uninformative_adapter_reads |
199835.0 |
| Pct_uninformative_adapter_reads |
1.9984 |
| Trimmed_reads |
9800165 |
| Trim_loss_rate |
2.0 |
| Peak_adapter_insertion_size |
34 |
| Degradation_ratio |
0.2305 |
| Aligned_reads_human_rDNA |
768600.0 |
| Alignment_rate_human_rDNA |
7.84 |
| Mapped_reads |
8644667 |
| QC_filtered_reads |
1123750 |
| Aligned_reads |
7520917 |
| Alignment_rate |
76.74 |
| Total_efficiency |
75.21 |
| Read_depth |
2.05 |
| Mitochondrial_reads |
267632 |
| Maximum_read_length |
100 |
| Genome_size |
3099922541 |
| NRF |
0.94 |
| PBC1 |
0.97 |
| PBC2 |
49.21 |
| Unmapped_reads |
386898 |
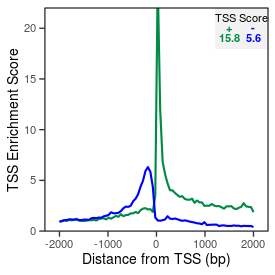
| TSS_coding_score |
15.9 |
| TSS_non-coding_score |
5.3 |
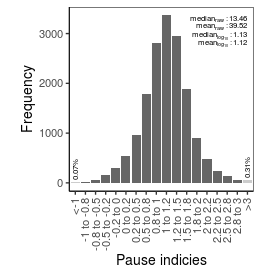
| Pause_index |
13.46 |
| Plus_FRiP |
0.36 |
| Minus_FRiP |
0.35 |
| mRNA_contamination |
2.63 |
| Time |
1:01:28 |
| Success |
02-27-10:24:31 |