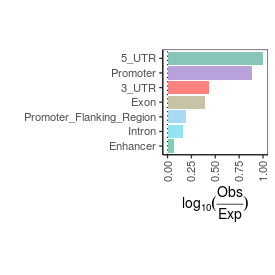
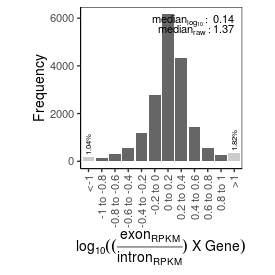
| sample_name |
K562_PRO-seq |
| sample_desc |
K562 PRO-seq |
| treatment |
none |
| replicate |
1 |
| toggle |
1 |
| protocol |
PRO |
| organism |
human |
| read_type |
SINGLE |
| cell_type |
K562 |
| purpose |
gold standard |
| umi_status |
FALSE |
| umi_length |
0 |
| data_source |
SRA |
| read1 |
/project/shefflab/data/sra_fastq/SRR1554311.fastq.gz /pro... |
| srr |
SRR155431[1-2] |
| srx |
SRX683602 |
| Sample_title |
K562 PRO-Seq |
| Sample_geo_accession |
GSM1480327 |
| Sample_source_name_ch1 |
K562 cells |
| Sample_characteristics_ch1 |
pull-down substrate: streptavidin |
| Sample_extract_protocol_ch1 |
PRO-seq libraries were prepared as described previously (... |
| Sample_description |
This Sample represents two sequencing replicates. |
| Sample_data_processing |
Supplementary_files_format_and_content: Processed data fi... |
| Sample_platform_id |
GPL16791 |
| Sample_contact_name |
Leighton,James,Core |
| Sample_contact_email |
ljc37@cornell.edu |
| Sample_contact_laboratory |
John T. Lis |
| Sample_contact_department |
Moleular Biology and Genetics |
| Sample_contact_institute |
Cornell University |
| Sample_contact_address |
417 Biotechnology Building |
| Sample_contact_city |
Ithaca |
| Sample_contact_zip/postal_code |
14853 |
| Sample_contact_country |
USA |
| Sample_instrument_model |
Illumina HiSeq 2500 |
| Sample_library_selection |
other |
| Sample_library_source |
transcriptomic |
| Sample_library_strategy |
OTHER |
| Sample_relation |
SRA: https://www.ncbi.nlm.nih.gov/sra?term |
| gsm_id |
GSM1480327 |
| Sample_supplementary_file_1 |
ftp://ftp.ncbi.nlm.nih.gov/geo/samples/GSM1480nnn/GSM1480... |
| Sample_supplementary_file_2 |
ftp://ftp.ncbi.nlm.nih.gov/geo/samples/GSM1480nnn/GSM1480... |
| Sample_series_id |
GSE60456 |
| File_mb |
38517.59 |
| Read_type |
single |
| Genome |
hg38 |
| Raw_reads |
496581677 |
| Fastq_reads |
496581677 |
| Reads_with_adapter |
423059183.0 |
| Uninformative_adapter_reads |
12437983.0 |
| Pct_uninformative_adapter_reads |
2.5047 |
| Trimmed_reads |
484143694 |
| Trim_loss_rate |
2.5 |
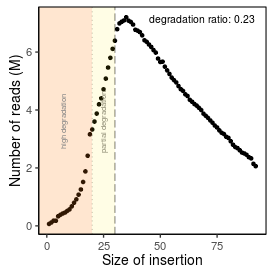
| Peak_adapter_insertion_size |
34 |
| Degradation_ratio |
0.2312 |
| Aligned_reads_human_rDNA |
44680364.0 |
| Alignment_rate_human_rDNA |
9.23 |
| Mapped_reads |
434069790 |
| QC_filtered_reads |
46741044 |
| Aligned_reads |
387328746 |
| Alignment_rate |
80.0 |
| Total_efficiency |
78.0 |
| Read_depth |
29.61 |
| Mitochondrial_reads |
9149071 |
| Maximum_read_length |
100 |
| Genome_size |
3099922541 |
| NRF |
0.28 |
| PBC1 |
0.54 |
| PBC2 |
3.89 |
| Unmapped_reads |
5393540 |
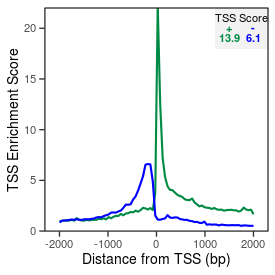
| TSS_coding_score |
13.9 |
| TSS_non-coding_score |
5.8 |
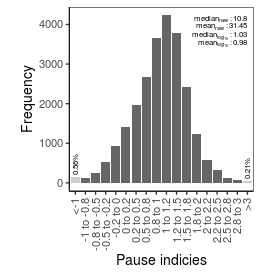
| Pause_index |
10.8 |
| Plus_FRiP |
0.36 |
| Minus_FRiP |
0.34 |
| mRNA_contamination |
1.37 |
| Time |
13:05:37 |
| Success |
02-27-22:28:40 |